Basic Usage Example
In this example we show the simplest usage of BoolXAI.RuleClassifier.
Input data
First, we need some binarized data to operate on. We’ll use the Breast Cancer Wisconsin (Diagnostic) Data Set, which can be loaded easily using sklearn:
[1]:
from sklearn import datasets
X, y = datasets.load_breast_cancer(return_X_y=True, as_frame=True)
# Inspect the data
print(X.shape)
X.head()
(569, 30)
[1]:
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | 0.07871 | ... | 25.38 | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 |
| 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | 0.05667 | ... | 24.99 | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 |
| 2 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | 0.05999 | ... | 23.57 | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 |
| 3 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | 0.09744 | ... | 14.91 | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 |
| 4 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | 0.05883 | ... | 22.54 | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 |
5 rows × 30 columns
Binarizing the data
All features are continuous, so we’ll use sklearn’s KBinsDiscretizer to binarize the data. Note that in general, we expect there to exist a better binarization scheme for most datasets - we use KBinsDiscretizer here only for pedagogical reasons. Unfortunately, KBinsDiscretizer does not return legible feature names. For this reason, below we use a patched version of KBinsDiscretizer which we call BoolXAIKBinsDiscretizer (located in util.py):
[2]:
# Transformers should output Pandas DataFrames
from sklearn import set_config
# Make sure that the transformer will return a Pandas DataFrame,
# so that the feature/column names are maintained.
set_config(transform_output="pandas")
# Binarize the data
from util import BoolXAIKBinsDiscretizer
binarizer = BoolXAIKBinsDiscretizer(
n_bins=10, strategy="quantile", encode="onehot-dense"
)
X_binarized = binarizer.fit_transform(X)
X_binarized.head()
print(X_binarized.shape)
X_binarized.head()
(569, 300)
[2]:
| [mean radius<10.26] | [10.26<=mean radius<11.366] | [11.366<=mean radius<12.012] | [12.012<=mean radius<12.726] | [12.726<=mean radius<13.37] | [13.37<=mean radius<14.058] | [14.058<=mean radius<15.056] | [15.056<=mean radius<17.068] | [17.068<=mean radius<19.53] | [mean radius>=19.53] | ... | [worst fractal dimension<0.0658] | [0.0658<=worst fractal dimension<0.0697] | [0.0697<=worst fractal dimension<0.0735] | [0.0735<=worst fractal dimension<0.0769] | [0.0769<=worst fractal dimension<0.08] | [0.08<=worst fractal dimension<0.0832] | [0.0832<=worst fractal dimension<0.089] | [0.089<=worst fractal dimension<0.0959] | [0.0959<=worst fractal dimension<0.1063] | [worst fractal dimension>=0.1063] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 3 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
5 rows × 300 columns
Training a rule classifier
We’ll now train a rule classifier using the default settings:
[3]:
from boolxai import BoolXAI
# Instantiate a BoolXAI rule classifier (for reproducibility, we set a seed)
rule_classifier = BoolXAI.RuleClassifier(random_state=43)
# Learn the best rule
rule_classifier.fit(X_binarized, y);
We can print the best rule found and the score it achieved:
[4]:
print(f"{rule_classifier.best_rule_}, {rule_classifier.best_score_:.2f}")
And(~237, ~209, ~259, ~238, ~236), 0.91
Now that we have a trained classifier, we can use it (and the underlying rule) to make predictions. As a quick check, we predict values for the training data, and then recalculate the score, using the default metric (balanced accuracy):
[5]:
from sklearn.metrics import balanced_accuracy_score
# Apply Rules
y_predict = rule_classifier.predict(X_binarized)
score = balanced_accuracy_score(y, y_predict)
print(f"{score=:.2f}")
score=0.91
We see that the score indeed matches the best score reported by the classifier, in the attribute best_score_. Note that one can use other metrics. However, it’s important to use the same metric for training, by passing it into the rule classifier using the metric argument, as is used for evaluation.
Making sense of the rules
The above rule uses the indices of the features which makes it compact, but it’s also hard to interpret. We can replace the indices with the actual feature names:
[6]:
rule = rule_classifier.best_rule_
feature_names = X_binarized.columns
rule.to_str(feature_names)
[6]:
'And(~[926.96<=worst area<1269.0], ~[worst radius>=23.682], ~[worst compactness>=0.4478], ~[1269.0<=worst area<1673.0], ~[781.18<=worst area<926.96])'
We can also plot the rule using the plot() method. Again, we have the option to pass in the feature names:
[7]:
rule.plot(feature_names)
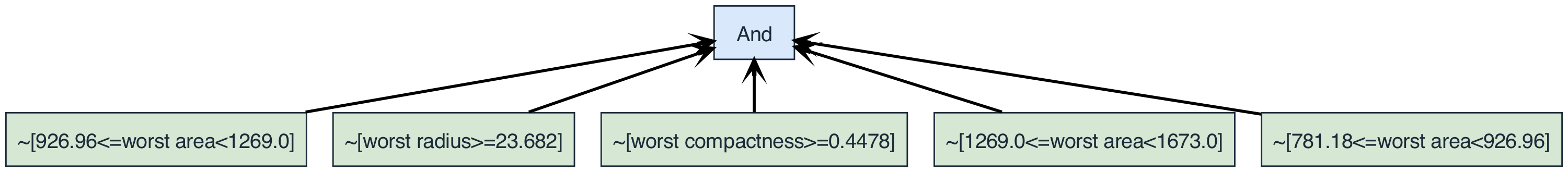
We define the complexity of a rule to be the total number of operators and literals. The depth of a rule is defined as the number of edges in the longest path from the root to any leaf/literal. We can inspect both for the above example:
[8]:
print(f"Complexity: {rule.complexity()}") # Note: len(rule) gives the same result
print(f"Depth: {rule.depth()}")
Complexity: 6
Depth: 1
For programmatic access it can be convenient to have a dictionary representation of a rule, optionally passing in the feature names:
[9]:
rule.to_dict(feature_names)
[9]:
{'And': ['~[926.96<=worst area<1269.0]',
'~[worst radius>=23.682]',
'~[worst compactness>=0.4478]',
'~[1269.0<=worst area<1673.0]',
'~[781.18<=worst area<926.96]']}
It’s also possible to get the graph representation of a rule as a NetworkX DiGraph, once again, optionally also passing in the feature names:
[10]:
G = rule.to_graph(feature_names)
print(G)
DiGraph with 6 nodes and 5 edges
We can inspect the data in each node:
[11]:
print(G.nodes(data=True))
[(5095761872, {'label': 'And', 'num_children': 5}), (5095762064, {'label': '~[926.96<=worst area<1269.0]', 'num_children': 0}), (5095761728, {'label': '~[worst radius>=23.682]', 'num_children': 0}), (5095760576, {'label': '~[worst compactness>=0.4478]', 'num_children': 0}), (5095760816, {'label': '~[1269.0<=worst area<1673.0]', 'num_children': 0}), (5095760864, {'label': '~[781.18<=worst area<926.96]', 'num_children': 0})]
The name of each node is set using id(), which returns the memory address of that object. This is done to make sure that the name of each node is unique. Each node contains two additional attributes. The label key contains the label of each node, and num_children is the number of children of that node. Literals have zero children (they are leaves), and operators must have two or more children.